Karen Marshall – forum organiser
-
-
-
-
-
-
Karen Marshall – forum organiser wrote a new post, Building a genomic reference resource on African cattle 5 years, 8 months ago
Here we will discuss the need and potential for a Genomics Reference Resource for African Cattle – which is a collated and publicly accessible set of sequence information on African cattle breeds. The sequence […]
-
Raphael Mrode replied 5 years, 5 months ago
In developed countries, individual countries have realized that they could not build adequate genomic reference resources on their due cost and the number of animals required. Consequently we have blocks of countries such as the North American Consortium consisting of USA, Canada, Italy and UK and the Euro-genetic consisting of France, Germany, Finland etc, exchanging genomic information. In small breeds such as the Brown Swiss, several countries pool their genomic information together to reduce cost and to achieve adequate numbers for meaningful genomic analysis. Therefore the building of an Africa genomic reference resource must cut across country bounders and strategic. Just as John Gibson remarked, we must move beyond mere characterization to actual implementation procedures that will have impart on productivity. Thus sampling of animals should be strategic; influential sires could be sampled so that we collect data from their daughters leading to genomic analysis that has practical application.
-
since it is termed ‘African’ then it should be at least representative of all the breeds with proven impact on the locals; interms of the performance & productivity. There are practically no substantial up-to-date records for these animals which means there may be a considerably reliance on the knowledge of the farmers to identify the influential sires and also track the progenies. The Establishment of a farmer research network may be helpful both for these strategic sampling as well as subsequent dissemination of the results for practical application and impact
-
Thanks – we will come back to the issue or breed prioritization later in the survey, as we will have to make some initial choices as the resource builds up
-
-
Yes I agree on strategic selection of animals – we can priorities influential animals when we have that information, but unfortunately their are few cases of this. It would be great if others could comment on breeding / recording programs on African cattle of which they are aware
-
I quite agree with the submission of Raph. One of the long-range objectives of the cattle Genome Program is to identify all coding sequences, genes and other functional elements in genomic DNA. Given that physical maps are being rapidly assembled and that the rate of accumulation of large-scale sequence data is increasing, there is a critical need for robust, high-throughput, and cost-effective methods and strategies to identify and map functional elements in the genome. An Indiana local dairy farmer claimed that DNA marker-technology has helped him boost production to 40 litres of milk per cow, per day, Although the 50K test continues to be used for high-end breeding stock and screening young artificial insemination (AI) sire entries, a simpler, cheaper test called the 3K test, which identifies about 3,000 carefully chosen SNPs, has been developed to allow herd owners to make use of genomic testing on a wider basis. Provided that at least one parent has been tested with the 50K test, the more economical 3K test can be used with only a small loss in accuracy due to the use of a method called imputation, which uses knowledge of the parental genome in the calculations. Recently, more SNPs and improved accuracy have been added to the 3K test, now called the 6K test. Due to the cost implication, I will recommend the use of 6K genomic application. The smallholder dairy farmers may be organized into a formidable group in such a way that they can pool resources together in order to be able to pay for the genomic services inform of public-private partnership. Such genomic services should be highly subsidized by the government in power so that the farmers will be able to afford them.
-
-
Peter Freeman replied 5 years, 5 months ago
Couldn’t agree more. Of course ‘advocacy’ shouldn’t be left to chance – we have to make a business case for WHY donors should get on board. Maybe think of them as ‘investors’ in tropical livelihoods
-
Peter Freeman replied 5 years, 5 months ago
Can we move from taking cursory looks to actually mapping (or making lists of) the existing Genbanks and databases? Who would be well placed to do that? Do they know about us and could we get them interested?
-
Mick Watson replied 5 years, 5 months ago
We have started a bit of this, we’ll be able to do more once recruitment has finished!
-
I think we can leverage on the initiatives of AU-IBAR and exploit the potential of ILRI-BecA to work out modalities for GenBanks and databases for African Livestock species. Through the strong financial backing and political will of African Nations and the support of international funding bodies, we may be able to make a headway.
-
-
Peter Freeman replied 5 years, 5 months ago
Might be good to take a post-funding look at existing evidence-based arguments, such as they are, and take it from there. At CTLGH we’re keen to tap into past and present PROJECTS and their knowledge artefacts, which tend to disappear from view once funding is secured and spent, The compelling arguments are locked up in (often eloquent) project proposals and reports that are tossed aside once they do their job of winning funding, and the focus turns to “research impact” of academic publications rather than real world impact outlined in the project proposals. We need to join up projects to show their collective impact, and knowledge gaps, and then point these out to investors.
-
Yes, and those of us involved in Research for Development projects, are being increasingly asked to provide evidence on project outcomes (or, as you put it, real world impact)
-
-
Mick Watson replied 5 years, 5 months ago
Genome reference sequences are of huge value to researchers. It would be good to know what challenges exist, if any, in terms of making genome sequence data usable for scientists in Africa.
-
Mick Watson replied 5 years, 5 months ago
I completely agree with this and many of us are working to ensure that phenotypic data are collected in standard ways that allows and enables downstream analysis
-
Mick Watson replied 5 years, 5 months ago
Do we know why some stakeholders failed to engage?
-
I think for this to be successful it has to be partly driven by the farmers/stakeholders themselves as they are the final users of the innovation. The hurdle will be to have a design that easily incorporate the smallholder farmers whose lives we want to improve via genomics tools & application. The benefits needs to be well articulated beyond the science of it.
-
-
Mick Watson replied 5 years, 5 months ago
I suspect “the devil is in the detail” – the current Hereford assembly has errors in it, and also there will be quite a lot of structural variation between cattle breeds in Africa and the Hereford reference. We are also working on difficult-to-assemble regions such as MHC
-
Mick Watson replied 5 years, 5 months ago
John, very interested in your comments on this! How do we maximise the translation of genomics into results that change lives?
-
Thanks for your insightful comment. Yes I agree that we need early results / applications to stimulate future investments. Could you give some examples of what you have in mind.
I will come back to the issue of which breeds, how many animals, what information etc. a bit later in the forum
-
And please don’t forget to take the survey – the ‘take survey’ button is on the top bar
-
Many people have said this resource will be useful, but there has been limited discussion on how it will be used, who will benefit from its use etc.
It would be great to build on the perceptive comment of John Gibson –essentially that the resource design must lead to exciting and useful results generated from the early investment, to provide incentives for future investments.
Thus I propose the next discussion topic as “What are the strategic research questions we can answer, or applications we can facilitate, with such a resource” (the definitions of which will feed into later discussions on resource design)
-
Baitsi Podisi replied 5 years, 5 months ago
With respect to Yakubu’s comment on the need for financial and political commitment, all is not gloom and doom as African Leaders have done a positive thing in adopting the Livestock Development Strategy for Africa (LiDeSA) (http://www.au-ibar.org/strategy-documents) which is new advocacy tool to mobilise support and investment in livestock in Africa. What remains is for the technocrats and other stakeholders at country levels to domesticate the strategy within their respective countries.
Baitsi
-
Thanks for sharing the link to this cornerstone document. Advocacy on investment in African Livestock is indeed critical.
-
-
Investment in building a genomics reference for African cattle and African Animal Genetic Resources in general is worthwhile. But what is new in that. The CBD signed since 1992 identified the importance of animal genetic resources management and conservation and stressed the need to help countries in need (technical and funds) to do so.
My concern is should Animal Genomics be the priority for African countries? The answer is NO. Africa Animal Genomics should be The World priority of course (Go back to CBD paragraphs).Animal Genetic Resources priority No1 in Africa should be going back to the basics (Farmers Breed Associations formation, suitable chain production establishments with small farmers playing key roles, suitable breeding strategies with their complete ingredients: identification, recording, evaluation , dissemination of adapted productive animals, and sustainable breeding and crossbreeding programs). -
Asma replied 5 years, 5 months ago
Funding Animal Genomics in Africa will not serve Africa in the first place. It will help more parties that sell equipments and lab needs. Africa needs first to build ilt capacities in training and education in animal breeding and genomics.
-
Thanks for raising the excellent point on capacity. I will raise capacity building needs as a specific point in the forum next week.
-
Dr. V. E. Olori replied 5 years, 5 months ago
I think these things are going on in the background gradually. For example I came aross a new company started recently; http://www.africanbio.com; which can become a key player both in terms of training and service provision. It is African. I beleive funding if properly channeled can speed up capacity building and development of key infrastructure locally.
-
-
Welcome to week two of this forum!
This week I wanted to concentrate on the design of the potential Genomics Reference Resource for African Cattle, but to do so we need to be more specific on the current discussion topic “What are the strategic research questions we can answer, or applications we can facilitate, with such a resource“, as this knowledge will inform the design. Please do contribute your thoughts on this apparently challenging question.
-
Africa needs to establish its Continental Animal Gene Bank. This will help Africa to have its own cattle genomics reference. Is it a dream? Let’s make it happen.
-
Sabrine replied 5 years, 5 months ago
Creation of an African Platform in molecular genetics is a major step toward African cattle reference in Genomics .
-
-
Baitsi Podisi replied 5 years, 5 months ago
The issue of an African genebank is already in the cards under the AU-IBAR animal genetics project. The planned genebank will be hosted the African Union Panafrican Veterinary Vaccine Centre (AU- PANVAC)
-
Thanks Baitsi – Great to hear of an African genbank. Which country is the AU-PANVAC in, and when is the genebank expected to be operational?
-
Baitsi Podisi replied 5 years, 5 months ago
The African Union Panafrican Veterinary Vaccine Centre (AU-PANVAC), provides vaccine quality control for African vaccine producing laboratories to meet international standards. The Centre promotes standardization and quality control of biological products and provides training and support services for vaccine production laboratories. It is based in Debre Zeit, Ethiopia.
-
-
-
Asma replied 5 years, 5 months ago
In establishing cattle reference using genomics, universities in Africa can play a key role if they are organized in a Network toward this objective
-
Yes – and National Agricultural Research Institutes should also be involved
-
-
In relation to our current discussion topic (applications of a reference resource) I would like to add a starting suggestion.
The use of genomic information to determine breed composition of admixed animals is being increasingly used (there were several examples of this in mentioned in this forum – most commonly in relation to performance comparison of different breeds / genotypes). However this requires the availability of reference information (genomic information on the underlying breeds present in the admixed animals) . If we had this reference information available and linked to a ‘smart tool’ to determine breed composition, researchers would no longer need to obtain the reference information on a project by project basis. Do others see utility in this?
-
Ben Abdallah replied 5 years, 5 months ago
Establishing Animal genetic resources programs including identification, recording and genetic evaluation and dissemination of adapted genes is an important step for Africa. This is the plus of phenomics.
-
monia replied 5 years, 5 months ago
For African Cattle reference in genomics,inventories should be done in all African countries to identify cattle breeds and populations
-
Sabrine replied 5 years, 5 months ago
Phenomics and genomics should go together in Africa to achieve comfortable levels in animal genetic resources management and conservation
-
monia replied 5 years, 5 months ago
Information generated by farm animals in Africa can help farmers better manage theirs animals and make better selection decisions if these infoemation are well processed. Africa Animal Genetic Ressources information can lead to a continent data bank useful to farmers and researchers. Empowering Africa Farmers toform breed Associations can be the first step to this goal
-
It seems that my photo disappeared just after I posted it. Only my name remained which is essential.
Using phenomics and genomics can mislead some of us. Genomics is ok to me, but what’s phenomics? What is behind phenomics?
Anyway, back to the subject and I will stick to what I think phenomics is or are! I read through the word phenomics, any science or technique linked to phenotypes of the animals including their performances. This is a big gap in Africa (at least the majority of African countries). There is an absence of complete and coherent animal breeding strategies similar to what we find in developed countries. I do not consider university or research institutions flocks or herds as representatives of what it should be done in our countries involving farmers with a majority of small farmers under low input systems. The challenge is here. The priority of national and international efforts, in my opinion should be at this level. Genomics should be a complementary component and not the alternative for Africa. This is why building capacities through training is essential, bringing farmers together to promote their native breeds is also a promising pathway for the future of Animal Genetic Resources in Africa. -
Asma replied 5 years, 5 months ago
Phenomics showed concrete results and progress in devloped countries. Through training and education, Africa can acheive a reliable building capacity in Phenomics to serve Africa Animal genetic ressources needs
-
Thanks Asma. I would like to see a lot more effort towards developing new phenomic approaches that will meet the needs of Africans systems. Note their is some more discussion on this taking place in the ‘discuss-using’ page.
-
-
Hi all – looking forward to more contributions moving into the last day of the forum (tomorrow – Friday 26th August). And also please don’t forget the survey (accessed by the ‘take survey’ tab above).
-
African animal genetic resources need to be worked on, the major challenge is our capacity to explore whether phenomics or genomics. mechanisms to build capacity first on phenomics and associate tools for evaluation is the primary thing
-
We come to the end of the forum time, and I very much thank everyone for their contributions. We will however leave the space open for the next few days, to allow for last comments and for people to view the summary posts.
Thanks Again, Karen.
-
Yes I believe building a gnomic resource reference for African cattle would be a worthwhile investment for the future. One key reason is for us to appreciate the value of our animals and most importantly target and utilize their unique traits of economic value in the development of appropriate cattle breeds for specific ecozones and production systems.
Specific research questions could be the level of introgression of exotic blood into crossbreds that will help ensure high productivity but at the same time not lead to the loss of adaptive and disease resistant genes. The type of breeds most suited to various production systems and climate smart breeds can also be targeted in our breeding programmes. Finally, in building the genomic reference resource for African cattle, many opportunities should open up for the training of more African cattle geneticists and building of networks between all stakeholders which are all key ingredients in the sustainable use of Africa’s cattle genetic resources.
-
-
Karen Marshall – forum organiser wrote a new post, Using genomic information on cattle in Africa 5 years, 8 months ago
Here we are discussing the applications of genomic (or genetic) information on cattle in Africa, both in the near future (next 5 years) and longer term.
Some potential applications are given below (see also […]
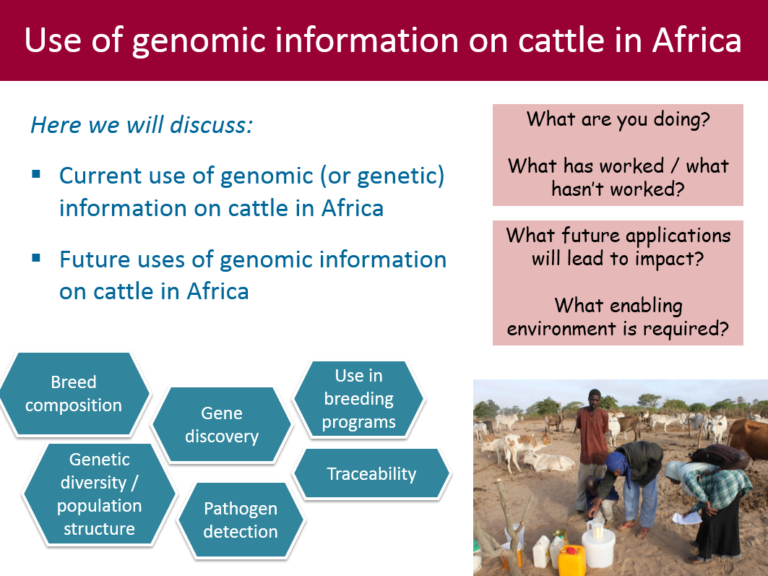
-
Mick Watson replied 5 years, 5 months ago
Hello! I think this is the kind of project and the kind of idea that all researchers interested in farm animal genomics would be interested in. There are two key questions: (i) what barriers exist to the genomic characterisation of Nigerian cattle and (ii) how do we translate the results into real impact for people?
-
Thank you Mick. Funding is the greatest obstacle. Lack of basic molecular equipment and facilities for cutting-edge researches is another while there is dearth of experts in the field of genetics and genomics. Also, inconsistency in livestock policies by successive governments have made some good projects to collapse midway. Good research products can only get to the farmers successfully if there is a well coordinated public-private partnership. However, at a small scale, research institutions through extension services could make research findings available to the farmers.
-
-
Mick Watson replied 5 years, 5 months ago
With the characterisation of pathogens you mention, did this lead to anything useful e.g. better vaccines or better intervention strategies? How do we improve our ability to tackle disease?
-
Yes, this has improved the vaccine production with actual impact as most of the vaccines being used for poultry & cattle are produced locally. A recommended minimum standards for bio security especially for diseases like ASF, AI has also being adopted.
-
-
Mick Watson replied 5 years, 5 months ago
I agree with this, so how do we do this? How do we engage with farmers and convince them of the need to engage with our efforts?
-
To this end, I would like to see a study on farmer incentives for engaging in recording schemes – we are working on the assumption that feedback to farmers (based on information they have submitted) that assists them in management decisions (health-care, feeding, breeding etc.) will provide this incentive, but that is yet to be well tested
-
-
Mick Watson replied 5 years, 5 months ago
I agree and I would be interested in your opinion on how best we could make such data available and usable by the African scientific community
-
Mick Watson replied 5 years, 5 months ago
Indeed – it’s very clear we need strong genotypic data alongside well defined phenotypic data from the same animals.
-
Mick Watson replied 5 years, 5 months ago
How do we influence the policy makers?
-
Mick Watson replied 5 years, 5 months ago
I am interested in your opinion about how best to make this type of data available and usable to African scientists
-
Mick Watson replied 5 years, 5 months ago
Thanks Kabuni. How do we engage with the farmers and get them on board? How do we convince them that the effort is worth it?
-
Kabuni Thomas replied 5 years, 5 months ago
Best farmers have to be selected and get involved in any breeding programme to represent the rest of the farmers, also these projects have to located on-farm where farmers feel a sense of belonging/ownership and will effectively participate and cooperate to implement the objectives of the programme. Therefore, once the programme is successful it will be easier for the other farmers to adopt the it.
-
-
Hi All. Thanks for the discussion to date on topic 1 – current applications of genetics or genomics to African livestock systems. We have collected a few examples of these, but please keep contributing. I will summaries all examples at the of the forum.
I would also like to add a new discussion topic – what genetic or genomic applications have the most potential to positively impact on the rural poor in Africa – in the near future and longer term?
-
Umar Aliyu Umar replied 5 years, 5 months ago
NAPRI/ ABU is having a programme for local farmers to improve milk production through crossing fresian to Bunaji or white fulani since late 1960s and the success is much to explain their quite a lot of publications in line with that. NAPRI has Beef research programme, dairy research programme and Animal reproduction and artificial insemination programme working together to desiminate technologies and breeding bulls for farmers in Nigera. Beef research programme has out station in Talata mafara at Zamfara state mainly for research using Sokoto gudali breed and it was reported to be the breed of choice for beef ( Madziga et al. , 2014 , shehu et al ., 2014 etc. ). Like wise white fulani was evaluated to be a dual purpose breed for milk and meat production. The dairy stock of NAPRI composed of Friesian-Bunaji and they are giving an excellent result ( 20L/ day).
-
Great to hear of these activities are the link or genetic improvement to dissemination programs.
-
-
And please don’t forget to participate in the survey – use the ‘take survey’ button on the top bar
-
Mizeck Chagunda replied 5 years, 5 months ago
Dear Karen and All. Firt thanks for this initiative and for moderating the discussion. Just like your example of the work you carried out in Senegal, we (Univeristy of Rwanda and SRUC of Scotland), are currently doing some work in Rwanda with smallholder dairy farmers through a PEARL project funded by BMGF. Due to lack of pedigree information in most cases, wewill use genomic approaches to assign breed-types to individuals. I strongly agree that we need to keep recording the phenotypes. I think that one thing we need to seriously think about on the continent is the aspect of stoping to consider individual projects as stand-alone activities. We need to start thinking of communities of projects and everything we collect some data or consider traits to measure, we should think of how the individual project data sets can contribute to the community without compomising the ability to address the individual projects’ specific objectives.
-
Excellent points, and great that genomic information will be used for breed composition also in Rwanda dairy
-
What is the underlying objective of the project – is it a breed comparison?
-
Mizeck Chagunda replied 5 years, 5 months ago
The underlying objective is not breed comparison per say. Its just part of it. We are working with farmers that received cows through the One Cow per Family Programme (Girinka). As you might be aware, in such programmes the focus is on passing on the first heifer to another farmer. One of our questions is, can we start identifying the bull calves as a source of germplasm and breeding lines? Given the different breeds and their crosses that are used and sparse pedigree data, genomic approaches will help us in this process. I hope this explains it in a nutshell.
-
-
-
Kabuni Thomas replied 5 years, 5 months ago
Among Animals’ Assisted Reproductive Technologies (AART) (eg. AI, MOET, Semen sexing, Gene cloning, etc.), AI stand to be the best way of applying genetic information due to its potentiality and positive impact to the poor rural African farmers. The uniqueness/specialities of AI as compared to other AART is that: The uptake of AI to farmers is simple and easy to practice, it is not very expensive, and easy availability and accessibility of AI technicians. However, for better performance AI programmes should include oestrus synchronisation protocols because of fertility problems (poor ovulation and irregular oestrus) in our herds which then will tend to maximize the reproductive performances of our cattle. To make the AI programme simple and more effective , FTAI (Fixed Time Artificial Insemination) can be used as a substitute of normal AI (Artificial Insemination) simply because in FTAI there is no need of detecting oestrus prior to insemination instead insemination is done based on fixed time. Under this regime farmers can be sure of increasing their productivity (i.e. increased pregnancy and conception rates, calving rates, increased weaning rates, and reducing calving intervals etc.) within their herds. Hence the livelihood of the farmers will be improved as well in general.
-
Thanks. We have a project here (ILRI) looking at scaling of fixed-time AI. Also, we are testing a recording and feedback tool for AI service providers that is mobile-phone based (and aimed at increasing AI success rate, as well as improving service provision)
-
-
Baitsi Podisi replied 5 years, 5 months ago
My comments are based on my past experience as a researcher in Botswana and my involvement in management of farm animal genetic resources at national and international levels.
My take is that genomic tools are not a silver bullet. Therefore for any livestock improvement effort to bear fruit requires that the basics need to be in place i.e. the conventional livestock recording systems which are not existent in most developing countries. Record keeping is quiet a challenge for most farmers. Tracking the mating is also a headache especially in communal grazing systems where bulls of different breeds can mate with cows with no control. The dairy sub-sector has a higher chance of making progress because of the ease of implementing recording and of managing breeding through A.I.
Otherwise within herd selection programmes in Government Research stations have limited chance of success due to no strategies of involving the communities to sustain the genetic progress e.g. in open nucleus set-up.
While A.I. is generally performs well in most settings for dairy & beef systems, MOET results remain very poor. Improving capacity for MOET is necessary for breed multiplication.
Given the high costs of past molecular characterization studies related to breed diversity in some developing countries. Such resources would have been better spent on phenotypic characterization to support development of breed standards and designing of breed selection programmes which are absent in most developing countries.More over because often such studies were undertaken with very limited number of markers due to budget constraints.
Current genomic tools offer a ray of hope but the basic infrastructure for conventional animal performance recording are still necessary.Baitsi
-
Yes excellent points.
I completely agree that well-designed, robust and sustainable breed improvement programs are absolute requirements (without that, genomic selection cannot add value), and that many issues need to be addressed to achieve this – institutional arrangements, farmer incentives, delivery systems etc.etc.
Given your experience, what are your thoughts on how we can encourage donors and stakeholders to invest in phenomic characterisation?
-
Kabuni Thomas replied 5 years, 5 months ago
Thanks Baitsi you have said it well, however to my opinion ineffective application of genomic tools is constrained by inefficient breeding programmes and policies in our African countries and that is why it is difficult for the implementers to disseminate genomic technologies to our farmers, because farmers have not been hold accountably and been bound by these programmes and policies. Therefore, in situation like this serious reformation has to be undertaken for the transformation of our African Livestock Production Systems.
-
-
Gilbert replied 5 years, 5 months ago
indeed Karen, unless farmers see the value of what they are recording for, their involvement in record keeping might be compromised. The technology ought to help these farmers see an immidiate value for their records.
-
Also, importantly, how farmers can improve their management practice based on record keeping. This, I feel, is likely to be a key incentive as it is immediate
-
Adamu Isa Mani replied 5 years, 5 months ago
Genomic selection will be difficult to implement in African continent because of the high cost of sequencing in addition to poor record keeping attitude of farmers. Worst still, where researches are conducted at stations by institutions, farmers are not usually involved, and this, makes it hard for the farmers to adopt whatever comes from the research stations. If farmers can be persuaded to keep records as well as involving them and their goals in researches conducted at stations, we may achieve something in that direction.
-
Yes in developed country dairy systems the use of genomic selection has reduced the cost of progeny testing. But do we have this evidence for African systems that are not analogous? I think more cost:benefit modelling studies would be useful to this end – particularly on the issue of what scale of operation is required.
-
-
Andrew Marete replied 5 years, 5 months ago
Hi,
Thanks for this constructive dialogue, I work on dairy genomics and bioinformatics in particular quantitative trait nucleotides affecting complex dairy traits and gene by gene interactions in the same. I suppose the main challenge in developing countries, as many have pointed out, is not only lack of proper performance recording, but routine uniform recording. This is key in reducing all the noise in the data and potentially increase the accuracy of prediction from analysis of such data. From my previously work at ILRI Nairobi, I recall a platform for dairy recording was in beta phase, am not sure if this platform is still in use. One solution would be to set up a central database where farmers send a FREE short text (from any cell) with performance recording data. This can either be interactive SMS or Structured SMS. In both instances, it involves a series of commands initiated by the farmer entering either his national ID/ farm id, then selects the animal(s) and records the performance and any other metadata deemed necessary. This eliminates data entry errors, and need for major data editing cost. The best traits to consider first would be production traits (moderate to high heritability, easy to record).
As pertaining to GS, there is need for a customized chip specifically for the Bos Indicus species, the current available SNP bead chips explain less than 45% of the variation of the tropical breeds. This means that even if one does GS, they will only be trying to capture less than 45% of the variation. This is very far from would be ideal since complex traits are influenced by many genes each with a small effect, as such, many genes that are not yet discovered in our populations would, probably, be overlooked. With the data already available in our research stations and partner institutions, and with partnership of SNP bead chip companies, we can easily develop a chip specifically for our breeds (or borrow a leaf from India who have developed a lot in the last decade in this field)
At the end of the day, we have to come up with solutions that incentivize the farmer to participate in such programs, such incentives could be Cow performance certificates from accredited breeding organizations and loans in form of cows, just to name but a few.-
Thanks Andrew. Note that in relation to paper-less data recording, it is possible to use new tools create for mobile devices – such as open data kit – to collect data along the lines you suggested. We have been testing this – along with provision of feedback to farmers based on their data.
-
A customised SNP-chip is one potential outcome of the genomics reference resource – further discussions on the potential genomics reference resource are taking place under ‘discuss building’
-
-
Kabuni Thomas replied 5 years, 5 months ago
l agree that we have a potential genetic resources to be exploited from our livestock although we have not effectively exploited enough these potentials for the improvement of livestock productivity and livelihood of the poor rural farmers. The inability of these failures is contributed by a number of factors but the most important ones include; animal factors (herd fertility problems, and inbreeding etc.) and human factors (inefficient breeding programmes and breeding policy). As already discussed previously, it should be careful noted that until when the underlying issues above are well tackled and properly solved is where we can now realise the best outcomes resulted from the application of genomic tools. The better performance produced from dairy cattle is because these animals have less fertility and inbreeding problems as compared to the beef cattle. Again much genomic works/researches/projects have been conducted using dairy cattle compared to beef cattle. Therefore, it is now the right time to intervene in the beef cattle industry which accounts high contribution to the livestock sector and improve the performance of beef cattle owned by majority of our pastoral and agro-pastoral farmers using genomic tools.
-
…. though at present there are few (if any) government institutions with sufficient animal numbers and recording capabilities to generate the needed data for reference populations
-
Hi all – looking forward to more contributions on current genomics / genetics work being performed on livestock systems in Africa, and what you think are the key applications of genomics / genetics that will impact positively on Africa’s rural poor. A summary of discussions to date on the first point can be found here.
-
Sahar Ahmed replied 5 years, 5 months ago
Dear All
It is important to establish an African genetic consortium for African cattle and other animals. This will help a lot for genetic improvement, especially most the African countries have the same environmental condition (adaptation of animals for environmental stress) , some countries have the same breeds. For animal resistance: in most African countries animals facing the disease (adaptation of animals for environmental stress)
-
-
Baitsi Podisi replied 5 years, 5 months ago
It depends on what type of data needs to be collected. For growth traits (live weights, birth, weaning, yearling etc) which involve animal handling requires equipment (weigh scales), infrastructure (crushes) and concerted human effort especially with beef animals which tend to become restless when handled. The cattle owners need to see the value of such tedious work which calls for farmer education and supporting legal framework for them to reap the benefits of producing performance tested animals. With credible bodies to accredit the quality of identified elite animals justifies the investment by farmers.
About 10yrs back we operated a pilot beef cattle performance recording and evaluation scheme in Botswana. We used mobile scales drawn behind a 4×4 van in sometimes sandy terrain and a requirement for a farmer to have a crush to participate in the scheme. Ultimately farmers were encouraged to buy their own scales or bear the inconvenience of sharing a mobile scale.
Cheaper tools such as weigh bands and the use of ICT tools to collect and send data offers a convenient option which is not readily harnessed due to low literacy among farm workers who are left to tend the animals in remote locations with low mobile phone network penetration. A proliferation of phone Apps offers a good opportunity to facilitate data collection. Provision of farmer training and timely feedback to farmers is paramount to get buy-in.
Baitsi
-
Great to hear about experiences from India!
Yes what you propose would be immensely beneficial. Can you say more on what India is doing towards building the required reference populations (with both genotyped and phenotyped animals) needed for this?
-
Hi – Thanks for your comment. Please expand on what you would like debated – as this is a great forum for that.
-
Yes and the record keeping could also inform farm management decisions – which I think is a more immediate incentive than monitoring genetic progress.
-
HI – yes I agree that there has been a lot of confusion related to the term superior – many people think of this only in the terms of e.g. milk yield or growth rate, rather than also adaptability to harsh environmental conditions, disease resistance etc. Breeding animals is suit the needs of the production system / environment they are in, is very key.
-
Yes I agree that increased productivity requires a ‘intervention package’ approach – jointly addressing genetics, feeding, and health-care amongst others. Also, that farmers need access to markets, a voice in value chain governance etc.
I would like to hear more from experts in genomic selection on their thoughts in relation to size of reference populations, and level of ongoing phenotypic recording.
-
Thanks Chanda. Africa also has the problem of scarcity of impact-oriented genomic studies, and it would be great to see more focus on this.
-
On the question “Which genomic or genetic applications have the most potential to positively impact on the rural poor in Africa – in the near future and the longer term” we have had a number of responses in relation to genomic selection, including the varied constraints associated with implementation of such a scheme. But what about other genomic applications (determination of breed composition, livestock product traceability, pathogen detection etc.) – do you see future demand for these in African (or other developing) livestock systems?
-
Dr. V. E. Olori replied 5 years, 5 months ago
I believe the biggest impact of genomics will be on performance improvement through geneomic selection. The present constraints should be addressed rather than be seen as obstacles to attainement of breed improvement through genomics. I say this because Genomics offer us the tools for a more precise breed improvement which is essential to maintain the balance between performance and adaptability of the indigenous breed use by most rural poor farmers.
-
-
Kabuni Thomas replied 5 years, 5 months ago
Yes Karen there is great need of making sure breed composition determination, livestock traceability, and pathogen detection are in place within our livestock production systems. This is because our livestock products are banned from entering overseas markets simply they lack these information and we therefore fail to fully exploit the international markets in most of the developed countries.
-
Thanks – do you know of any countries actively working on traceability?
-
Kabuni Thomas replied 5 years, 5 months ago
Currently l have not come across with any African country with animal traceability programme which is in place working. For instance in Tanzania we have animal traceability policy but it is not yet practically well implemented and this is the problem of most of the African countries facing at the moment.
-
Thanks for the update Baitsi. Do you see any demand for tracing of livestock products (where genomics could play a role) versus tracing animals (where ear-tags seem more cost-effective)?
-
I only know of the two reports commissioned by AU-IBAR which myself and others helped in validating (TCP/RAF/3403 FAO/AU-IBAR) on animal genetic resources management in Central and West Africa (1. Support to pastoralism for the management and use of AnGRs in West and Central Africa and 2. Livestock identification and recording in Central and West Africa and impact on regional trade).
-
-
I concur with the submission of Kabuni. If we have to make the best use of our natural endowment, then every aspect of livestock breed composition, production, improvement, traceability, health and marketing must be put into consideration.
-
-
Several people have mentioned using genomic approaches to determine breed composition of animals (where it cannot be assigned based on phenotype, or due to lack of recorded pedigree). Are their other examples of people using this in their research, or who would find this beneficial in relation to future research? And what about industry application?
-
And on parentage testing – many people who have completed the survey indicated that they felt this was in demand from stakeholders. Are there examples of livestock parentage testing in African systems, and if so for what purpose …. or how do you see this being beneficial (or not).
-
Raphael Mrode replied 5 years, 5 months ago
Firstly, I will give a summary of the some of the work done using genomics based on the Kenya data generated from the Dairy Genetic EAST Africa Project (DGEA). Just over a thousand cows (crossbreeds) were genotyped and test day milk records were also collected over a period of time. We were able to successfully determine the breed composition of these cows using admixture analysis. Secondly, were able to compute the genomic relationship matrix and undertake genomic prediction using GBLUP and BayesC. The important outcome was genomic prediction for several type of crosses could be implemented with accuracy varying from 30 to 40%. The results could be use to select a team of young bulls based on their genetic merit for use by farmers
So I see potential use of genomics in parent verification in future and hence less need to accurate pedigree recording and traceability (already mentioned in the discussion). In a current study, one of our students has discovered same dairy sires have been used in Kenya, South Africa and Zimbabwe. The genotyping of such sires used across several countries offer a great potential for an across country genomic evaluation resulting in more accurate team of young bulls. Genomics will offer opportunity to identify genome regions associated with various adaptive features in many indigenous African breeds and how these could be used in practical breeding program either using genomic selection plus gene editing or some sorted of modified marker-assisted selection.-
Thanks Raphael for this update of the Dairy Genetics East Africa project, as well as your thoughts on how this could expand in the future. Their is often discussion on what is the minimum size of a reference population needed for genomic selection in African systems, such as smallhold systems (where their is often a lot of environmental noise and thus lower trait heritabilities compared to intensified systems). What are your recommendations here.
Also, how did you handle the presumably small contemporary group size in this analysis?
-
-
Raphael Mrode replied 5 years, 5 months ago
I just want to comment on the point made by Charles in terms of basing GS on data from government Institutions and few producers initially. Actually this is a very good approach and one may regard reference population from government institutions and few producers as a nucleus herd of some sort where more detailed phenotyping could be implemented as well. However, if these reference population animals are managed differently from the small holder systems, then we may have an issue of GxE to address. Therefore it will be necessary a deliberate attempt be made to include records from the small holder system. Actually we are developing a proposal for goat improvement based on this concept in some Africa countries. If you are interested in being a part, let me know
-
Hello Raph! Please I am highly interested in your proposed goat project. Please do not forget to include Nigeria.
-
Umar Aliyu Umar replied 5 years, 5 months ago
I joined the appeal to include Nigeria
-
-
-
As pointed out by Karen in of her recent messages, there are many opportunities that genomic applications could afford the African livestock sector. Meanwhile, the key question for me is: how do we tackle the issue of animal recording and identification in the commonly practiced traditional livestock farming system of this continent? In as much as the extensive system still prevails in many African societies, maximizing the potentials of genomic or genetic applications is doubtful especially in the nearest future term.
-
Thanks Olawale. My thoughts are that we shouldn’t think of any intervention as something that is universally needed – we should target it to systems where demand for the intervention exist, and also where other ‘success factors’ apply (e.g. favourable policy and institutional environment, or ability to create these). My feeling is that recording and feedback schemes currently have higher demand in the intensifying systems compared to other systems, as in the intensifying systems their is more emphasis on keeping livestock for household income purposes, as well as market orientation. I mentioned earlier in the forum mobile phone based recording and feedback schemes we (ILRI) are testing, based on the premise that information provided back to the farmer that assists them with farm management decisions will provide the ongoing incentive for farmers to record. This model, however, is still being tested. I would be interested to hear from others their thoughts on this important topic.
-
-
It will be interesting to identify with precision any Genomics success story or positive Genomic actions in Africa and find out who are the pioneers (local scientists vs Hired Experts), (National funding vs Outside funding…projects…) and check their sustainability and impact on real farmers income.
-
Yes I agree, and if any forum participants have such examples please share them.
-
-
Welcome to week two of this forum – thank-you very much to those who have participated to date, and I look forward to more challenging discussion.
Many people have commented on the need for phenotype information on African livestock – both in its own right and in support of genomic applications (such as genomic assisted prediction of breeding values). In relation to this, I would like to propose the next discussion topic as “Is phenomics – the ability to measure livestock performance – a bigger challenge than genomics in African livestock systems and, if so, how can we increase phenomic capabilities“
-
Phenomics is certainly a bigger challenge and I don’t think it is limited to only the African Livestock systems although it could be much more challenging than others. Although Phenomics presents greater opportunity for livestock improvement however addressing the genomics capabilities first will be a good rung to start. Its difficult learning to run when we haven’t started crawling.
-
-
Ben Abdallah replied 5 years, 5 months ago
In order to establish à cattle references in Genemics for Africa, we should build in the existing by identifying African Institutions that are in advance in animal genomics
-
It would also be interesting to hear of examples of new phenomic approaches being used in African systems, as well as elsewhere.
-
Certainly, we cannot talk about genomics in isolation. According to a recent report, the demand for high throughput phenotyping may stimulate a migration from conventional laboratory to web-based assessment of performance and behaviour, and this offers the promise of dynamic phenotyping-the iterative refinement of phenotype assays based on prior genotype-phenotype associations. In this wise, we may have to generate the phenotypes from animals which may vary from one country to another. Such phenotypes, although may take some years to generate when standardized Ab initio will be more tenable than relying on specific information without enough validation. We may borrow a leaf from the current ACGG project being led by ILRI, Ethiopia where provision is made for both phenotyping and genotyping of chicken being tested. Also, we may align with the regional initiatives of AU-IBAR on the characterization and improvement of African Livestock. It is only when we have a common goal and pull resources together that we can make a reasonable progress.
-
Umar Aliyu Umar replied 5 years, 5 months ago
Really appreciate the forum even though my contribution wasn’t much due to bad network in the area I’m living. In a nut shel we need an innovation platform for the African cattle and the issue of gene transfer should not be in place our animals has the potentials to do what ever we want and can be achieved through genomic selection as discussed previously.
-
-
-
Heather Burrow replied 5 years, 5 months ago
Thanks for your efforts in moderating this forum Karen. I have relatively recently undertaken two separate literature reviews (for different purposes, one a conference presentation covering beef & dairy cattle, sheep & goats grazed in extensive production systems in the tropics and the second specifically focused on the genetics of adaptive traits in beef and dairy cattle). In both reviews the major conclusion was along the lines of the following .. ”The greatest limitation to genetic improvement of these [productive and adaptive] traits for the foreseeable future is likely to be the lack of accurate phenotypes on which selection can be based. Development of new technologies is now offering some potential for new approaches to overcome this limitation.’
I am therefore currently working with a wide group of researchers (from the USA, Canada, UK, South Africa, Brazil & Australia not in any order of importance!) to develop a funding proposal specifically focused on alternative approaches to cost-effectively and practically measuring resistance/tolerance of cattle to both cattle and multi-host tick species, with the aim of approaching several funding agencies to co-invest in a new project that would evaluate several alternative methods and comparing the alternative approaches to the current but infeasible ‘gold standard’ of tick counts. We have the potential use of two well-equipped research stations and relevant cattle herds + experienced field technicians and research station managers (one each in South Africa and Brazil, with the possibilities of undertaking artificial tick infestations or using natural infestations if that is a preferred experimental approach) and an offer by one of the world’s leading genomic statisticians to design the experimental populations with the aim of using the results in nucleus cattle populations that have been and are continuing to be established with the specific purpose of improving all traits in the relevant breeding objective using genomic selection approaches.
The primary aim of the proposed research will aim to deliver benefits to Africa, South America and Australia, with spin-off benefits to other countries also impacted by ticks and tick-borne diseases (latest estimates being that ~80% of the world’s cattle are at risk from ticks & tick-borne diseases with losses estimated to be of the order of US$20-30 billion p.a.). Our thinking is that development of a ‘model’ proposal based on a co-investment approach for tick resistance in beef (and dairy) cattle might serve as the basis of a similar funding model for use in developing new phenotypes for other (particularly adaptive) traits in cattle and other grazing and intensively reared livestock. As well, the new technologies being trialed to assess tick resistance in cattle might also have value for use in testing other traits in cattle and other livestock species.
I would be happy to add anyone interested in this approach to a new mailing list that will be set up over coming weeks …
-
Thanks Heather – this is an extremely exciting initiative!
-
-
Ben Abdallah replied 5 years, 5 months ago
Establishing Animal genetic resources programs including identification, recording and genetic evaluation and dissemination of adapted genes is an important step for Africa. This is the plus of phenomics.
-
Thanks Okeno – your comments align with what many others have contributed to the forum. Great to see a general consensus.
-
Dr. V. E. Olori replied 5 years, 5 months ago
I believe phenomics is a big challenge and genomics without first sorting out the phenomics issue will be a bigger challenge. Recording phenotypic data is a challenge because there is no concerted effort towards improvement of any trait hence need for recording the trait. Where such interest exist, then the challenge is a structure that will allow the highly dispersed farmers to be enrolled in a recording scheme. First you need to convince the farmers why such regular data collection is important or beneficial to them. Then you talk about the cost, methodology, personel and tools of doing the actual recording.
So the challenge, as I see it, is the of absence an organisational structure resulting from a motivation to bring about change. If we can set up the institutions to facilitate this, African farmers will be open to data recording which will kick start the livestock improvement process. Genomics can then follow naturally. We need an institution or institutions to champion change and manage the whole process of livestock improvement at national and perhaps regional levels in Africa.-
Thanks Olori for your insights. We had some earlier discussion on incentives for farmer data recording – do you have any additional suggestions here?
-
-
Hi all – looking forward to more contributions moving into the last day of the forum (tomorrow – Friday 26th August). And also please don’t forget the survey (accessed by the ‘take survey’ tab above).
-
Nerry Corr replied 5 years, 5 months ago
Dear All, I have been late to join this discussion mainly because am in an area where the network connections are not good, it has been very interesting, i took time yesterday and today to go through almost all the discussions and i commend Karren for initiating such a forum. Phenomics as it is now is a real challenge, however i would want to suggest that African governments be put to a challenge to develop livestock policies that will incorporate tracability and record keeping for all livestock, this is not easy in countries where the level of illiteracy is high, but such programs adult literacy can be incorporated in the extension programs for livestock farmers. we developedd a recording method with rural farmers while disseminating elite bulls from ITC for multiplication by farmers, the farmers were using their family members who are going to school to do the recordings, I think if such programs are introduced to farmers they will notice the importance of animal record keping.
-
-
Tesfaye replied 5 years, 5 months ago
I wanted to share research in Ethiopia on estimation of breed composition level using Ancestory Informative MArkers (AIMs). Recent results showed few AIMs (~ 50 SNPs) accurately estimated the Awassi level in the Awassi x local crossbred population in the highlands of Ethiopia. We used this information to reccomened different level of Awassi introgression for different areas after associating the genomic information with the performances of the sheep.
Once the AIM SNP pannel developed, cost to estimate the breed composition of an animal is cheap (price to genotype a SNP is in the range of 3 to 10 euro cents). I guess this would be highly attractive and applicable in developing countries especially in dairy sector. For example in Ethiopia, due to poor or absence of recording and indiscriminate crossbreeding level of introgression of Holestein in to local Zebu breed is uknown. farmers and investors engaged in dairy business are highly curios to know the breed comopsition of the animal the are using. In addition, knowing the level of breed composition is mandatory to implement well structured crossbreeding. Ethiopian Biodiversity Institute has developed a proposal to use this tool to quantify the level of admixture of exotic blood in the dairy cattle population.
The use of high density marker for breed characterization and for detection of selection signature might also be another area of interest.
So my suggestion is to to strengethen and start use genomic tools for breed composition, deversity studies and detection of selection signature. At the same time establishing national pedigree and performance recording is crucial. These will lead us to use genomic tools for genomic selection and association studies.
-
Thanks – and this aligns with some of the earlier contributions on current and potential use of genomics. In particular, one of the most commonly discussed applications was around determination of breed composition.
-
-
Isidore Houaga replied 5 years, 5 months ago
Phenomics is actually a challenge in African Livestock breeding system. However, it would be better to associate the phenomics to the genomics rather than trying to sort out first the Phenomics issue. There is lack of genetic associative study in African indigenous livestock and this is unfortunate for development of a sustainable breeding programme.
-
In agreement with most contributors (especially Dr. Okeno Otieno & Dr. V.E. Olori), phenomics is a bigger challenge in African livestock system. Efforts should be channeled more towards establishing phenomics and well defined conventional breeding schemes. No doubt genomics is great as it facilitates rapid genetic improvement, but this is not possible without phenotypes. Even in developed countries where genomics is currently implemented, there is still emphasis on the collection of more and more phenotypes. That goes to show how important phenotypes are. As Prof. Mike Coffey of University of Edinburgh rightly puts it “In the era of genomics, phenotype is king”. So, be it for genome-wide association studies which enables us to link phenotypes to genes or genomic selection which could lead to rapid genetic improvement in African livestock system, phenomics is paramount.
-
The current state of livestock production in Africa is pathetic despite the huge potential of these animals to sustain the livelihoods of farmers. If phenomics and genomics could help improve livestock production and productivity, then African countries should not put these at the back burner. We should move away from our current crude ways of doing things by exploiting modern technologies that will contribute to increased livestock populations and more income to the farmers. Although the task is daunting, I believe if all the relevant stakeholders (governments, research institutions, farmers, regional and sub-regional organizations, private investors, NGOs and funding bodies) are highly committed and give phenomics and genomics the much deserved priority, then we will overcome. All current efforts geared towards improving African livestock especially cattle should be harmonized to yield laudable results. However, we should not forget the aspect of capacity building as it affects researchers and farmers. On a final note, I wish to commend the organizers of this e-discussion for a job well executed. Please keep the good work up.
-
Mizeck Chagunda replied 5 years, 5 months ago
One challenges with phenomics (as applied in functional genomics) is not phenomics itself. Its much more to do with how we train the next generation of scientists. Correct me if I am wrong but I see a lot of lack of connectedness in the way most courses are taught. Take an example of biochemistry, mathematics, biology and animal science (including animal breeding, animal health and animal nutrition). Until when the teaching/learning shows serious connections, the application of the omics in our production systems will only look good on paper but not on the ground where the animals are.
-
Thanks MIzeck and I fully agree – integration across disciplines is essential!
-
-
We come to the end of the forum time, and I very much thank everyone for their contributions. We will however leave the space open for the next few days, to allow for last comments and for people to view the summary posts.
Thanks Again, Karen.
-
lubna replied 5 years, 5 months ago
Dear all. Sorry for delay. Here in Sudan we have some animals are indangered. I search in one of them (nubian ibex) I would like to study the genetic diversity/population structure and assess the gene intoregression between ibex and do mastic goats in red sea state in Sudan.
i would like to do share the samples of DNA with most countries where ibex is exist.
I am looking forward to a fruitful collaboration -
NGONO EMA replied 5 years, 5 months ago
I will just join those who believe that we first have to ensure phenomics in Africa before moving forward to genomics.How is it possible to have for example the breeding values of our cattle breeds when there is no recording of their performances?We have to convince farmers that it is very important to keep consistent records on the performances of their animals.Of course prior to that phenotypic identification of our breeds is a must.On my own point of view we need to first focus on this.
-
-









